Journal of Microbiology & Microbial Technology
Download PDF
Review Article
Microbes - The Key Players in Anaerobic Digestion for Biogas Production
Ukaegbu-Obi KM* and Ifeanyi VO, Eze VC
Department of Microbiology, College of Natural Science, Michael
Okpara University of Agriculture, Abia State, Nigeria
*Address for Correspondence:
Ukaegbu-Obi KM, Department of Microbiology, College of Natural
Science, Michael Okpara University of Agriculture, Abia State,
Nigeria; E-mail: ukaegbu-obi.kelechi@mouau.edu.ng
Submission: 16 April, 2022
Accepted: 20 May, 2022
Published: 23 May, 2022
Copyright: © 2022 Ukaegbu-Obi KM, et al. This is an open access
article distributed under the Creative Commons Attribution License,
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Abstract
Microorganisms remain the powerhouse in the different processes
of anaerobic digestion (AD). These organisms in a synergistic
approach convert organic matters to biogas and other useful
products. Over time, the study of these microbes involved in AD has
been a challenge. This is because of the high limitations of the culturebased
techniques in the isolation of microorganisms. The advent of
molecular techniques in the study of microbes in AD has become a
groundbreaking achievement. Meta-omics give very deep insights not
only into the diversity of microbiota present but into the microbiome
and pathways involved. More studies are required using molecular
techniques to unlock more information about these tiny but powerful
living machines that help in balancing and maintaining both the
aerobic and anaerobic ecosystems.
Keywords
Anaerobic digestion; Biogas; Microbes; Molecular
techniques; Meta-omics
Introduction
The production of Biogas using anaerobic digestion has been in
existence but the role of microbes was known in the 20th century [1].
Various microorganisms with specific properties take part in the AD
process [2]. Microbiology of anaerobic transformation of organic
wastes is a process that involves many different groups of bacteria
in four main steps namely hydrolysis, acidogenesis, acetogenesis and
methanogenesis [2-8].
The individual degradation steps are carried out by different
consortia of microorganisms [4]. These organisms partly stand in
syntrophic interrelation and place different requirements on the
environment and in the final stage produce carbon dioxide and
methane, as the main products of the digestion process [9-11]. Besides
energy production, the degradation of organic waste also offers
some other advantages including the reduction of odour release and
decreased level of pathogens. Moreover, the nutrient-rich digested
residue could be used as organic fertilizer for arable land instead
of mineral fertilizer, as well as an organic substrate for greenhouse
cultivation.
The Bioprocesses of Anaerobic Digestion
Anaerobic digestion is widely adopted in the world, but the
microbial ecology of this process is still being studied [13]. Unfolding
and proper understanding of the complex structural diversity are very
important in understanding the functional relationship between the
various metabolic groups of microorganisms (hydrolytic, acidogenic,
acetogenic and methanogenic). Understanding this synergy will help
improve and optimize the process of AD thereby making it more
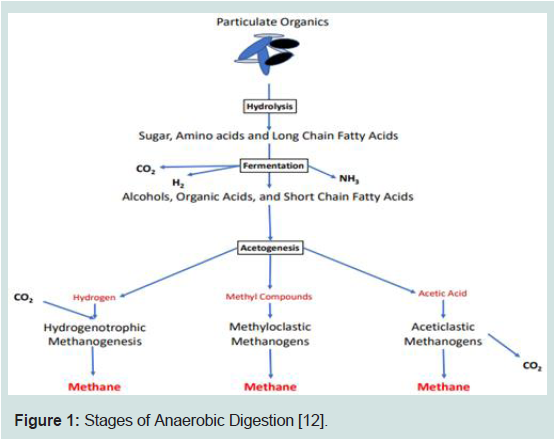
effective [13-14] (Figure 1).
Figure 1: Stages of Anaerobic Digestion [12].
Hydrolysis:
Complex organic molecules like proteins, polysaccharides, and fat are converted into simpler ones or soluble monomers like peptides,
saccharides, and fatty acids by exoenzymes like cellulase, protease,
and lipase produced by hydrolytic and fermentative bacteria [13,15-17]. The enzymes required for hydrolysis can either be attached to
microbial cells or secreted into the solution [18].Hydrolysis is a relatively slow step and it can limit the rate of
the overall anaerobic digestion process, especially when using solid
waste as the substrate [13,18]. The rate of the hydrolysis process
depends on such parameters as size of particles, pH, Production of
enzymes, diffusion and adsorption of enzymes on the particles of
wastes subjected to the digestion process. Different compounds have
different hydrolysis rates [19].
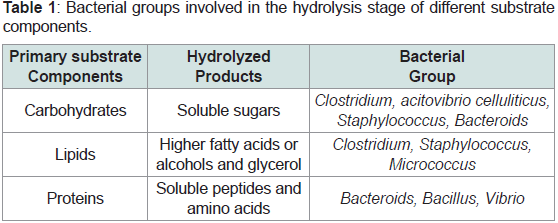
Several groups of hydrolytic microorganisms (strict anaerobes
and facultative anaerobes) [20] are involved in the degradation of
several substrate compositions, where the bacteria Bacteroides,
Clostridium and Staphylococcus are significant drivers [21] others are
Streptococcus, Enterobacterium [17,21]. Hydrolytic bacteria in AD are found within five phyla: Firmicutes, Bacteroidetes, Fibrobacter,
Spirochaetes, and Thermotogae [22]. Firmicutes and Bacteroidetes
are typically the most abundant taxa of hydrolytic bacteria in AD,
although the relative abundance of these taxa is often dictated by
inoculum and reactor type, as reviewed by [22] (Table 1) (2015).
In order to improve hydrolysis and anaerobic digestion
performance, several pretreatments (thermal, thermochemical
ultrasonic alkaline) have been carried out, which cause the lysis
or disintegration of sludge cells [23]. Hydrolytic bacteria can be
inhibited by elevated levels of Volatile Fatty Acids and hydrogen
partial pressure [22]. Acid accumulation and the process pH decrease
usually occur when hydrolysis occurs too rapidly, and this inhibits
methanogens [19].
Acidogenesis:
Hydrolysis is immediately followed by the acid-forming step -
acidogenesis [15].In the acidogenesis stage, acidogenic bacteria such as Lactobacillus,
Streptococcus, Clostridium etc transform hydrolysis products (amino
acids and sugars) into volatile fatty acids (VFAs) (acetic acid, butyric
acid, and propionic acid), organic acids (succinic acid and lactic
acid), ammonia (NH3), hydrogen gas (H2), carbon dioxide (CO2),
hydrogen sulphide (H2S), and low alcohols [16,17].
The higher organic acids are subsequently transferred to acetic
acid and hydrogen by acetogenic bacteria. It is always not possible to
draw a clear distinction between acidogenic and acetogenic reactions.
Acetate and hydrogen are produced during acidification and
acetogenic reactions and both of them are substrates of methanogenic
bacteria. The acidogenic and acetogenic bacteria belong to a large and
diverse group that includes both facultative and obligate anaerobes.
Facultative organisms are able to live in both aerobic and anaerobic
environments. Acidogenic facultative anaerobes present make use of
the oxygen that may be introduced into the digester during feeding
[24]. This action is very important in creating favourable conditions
for the obligate anaerobes.
As opposed to other stages, acidogenesis is generally believed
to proceed at a faster rate than all other processes of anaerobic
digestion, with acidogenic bacteria having a regeneration time of
fewer than 36 hours. With the rapidity of this stage, the production
of volatile fatty acids creates direct precursors for the final stage of
methanogenesis; VFA acidification is widely reported to be a cause
of digester failure [25]. During acidogenesis, the pH reduces [26].
When the acidogenesis rate is too high and the pH drop is significant,
severe inhibition of methanogens (methane-forming bacteria) will be
triggered [19]. The VFA other than acetate as well as some alcohols
is subsequently oxidized by syntrophic bacteria to acetate, hydrogen
and carbon dioxide.
Species that have been isolated from anaerobic digesters
include Clostridium, Peptococcus, Bifidobacterium, Desulfovibrio,
Corynebacterium, Lactobacillus, Actinomyces, Staphylococcus,
Streptococcus, Micrococcus, Bacillus, Pseudomonas, Selemonas,
Veillonella, Sarcina, Desulfobacter, Desulfomonas and Escherichia
coli [27]. These microorganisms are able to withstand low pH, high
temperatures and a high organic loading rate.
Acetogenesis:
During the third stage, namely acetogenesis, the VFAs, especially
acetic acids and butyric acids, are converted into acetate, H2 and CO2.
Among the VFAs (acetic acid, butyric acid, and propionic acid), 65-
95% of methane is directly produced from acetic acid [16]. As the end
product of these two processes - acidogenesis and acetogenesis is acid,
some researchers merge these two processes as acidogenesis [19].An obligate, syntrophic relationship exists between the acetogens
and methanogens. Syntrophy is the phenomenon in which one
species lives off the products of another species. About 64% of the
methane produced during anaerobic digestion comes from acetate,
while the remaining 36% comes from hydrogen [18].
Acetogenesis refers to the synthesis of acetate, which includes
the formation of acetate by the reduction of Carbon dioxide and
the formation of acetate from organic acids. Hydrogen-utilizing
acetogens, previously also termed homoacetogens, are strict anaerobic
bacteria that can use the acetyl-CoA pathway as (i) their predominant
mechanism for the reductive synthesis of acetyl-CoA from CO2, (ii)
terminal electron-accepting, energy-conserving process, and (iii)
mechanism for the synthesis of cell carbon from CO2 [28]. These
bacteria compete with methanogens for substrates like hydrogen,
formate, and methanol.
Organic acids (such as propionate and butyrate) and alcohols
(such as ethanol) produced during the fermentation step are oxidized
to acetate by hydrogen-producing acetogens. Electrons produced from
this oxidation reaction are transferred to protons (H+) to produce
hydrogen or bicarbonate to generate formate [29]. The production
of hydrogen exerts toxic effects on the acetogenic bacteria [20].
Collaboration with the hydrogen-consuming (hydrogenotrophic)
methanogens becomes essential, in a symbiotic process described as
syntrophy, in which the methanogens constantly utilize hydrogen to
produce methane [20].
Acetogens that oxidize organic acids obligately use hydrogen ions
and carbon dioxide as electron acceptors. The acetogenic bacteria
grow faster than the methanogens and the interaction of the two
groups of bacteria is important for the performance of the anaerobic
digester [28].
The coupling or syntrophic relationship of hydrogen producers
and hydrogen consumers is called interspecies hydrogen transfer.
Obligately syntrophic communities of acetogenic bacteria and
methanogenic archaea have several unique features: (i) they degrade
fatty acids coupled to growth, while neither the methanogen
nor the acetogen alone is able to degrade these compounds,
(ii) intermicrobial distances between acetogens and hydrogenscavenging
microorganisms influence specific growth rates [29], and
(iii) the communities have evolved biochemical mechanisms that
allow sharing of chemical energy. Typical homoacetogenic bacteria are Acetobacterium woodii and Clostridium aceticum [21]. Bacteria
that form the acetate by using butyrates and propionates respectively
are known as Syntrophobacter wolinii, Smithella propionica
and Pelotomaculum schinkii. Clostridium aceticum is another
microorganism that develops H2 and CO2 acetate. The accumulation
of hydrogen can inhibit the metabolism of acetogenic bacteria [30].
The maintenance of an extremely low partial pressure of hydrogen
is, therefore, essential for the acetogenic and Hydrogen-producing
bacteria [21].
Methanogenesis:
In general and as mentioned earlier, the Anaerobic Digestion of
organic material requires the combined activity of several different
groups of microorganisms with different metabolic capacities [30-32].
To obtain a stable biogas process, all the conversion steps involved in
the degradation of organic matters and the microorganisms carrying
out these steps must work in a synchronized manner.Methanogenesis is the last stage where methane and Carbon
dioxide are derived from acetogenesis products (acetic acid, H2,
CO2 and formate and methanol, methylamine or dimethyl sulfide)
by methanogenic bacteria [5,30]. It is a critical step in the entire
anaerobic digestion process, and its biochemical reactions are the
slowest in comparison to those in other steps.
Methanogens have longer duplication times (of up to 30 d)
and are generally considered the most sensitive group to process
disturbances [33]. The methanogens are the dominant species and
are strict anaerobes. They are vulnerable to even small amounts of
oxygen. They also require a lower redox potential for growth than
most other anaerobic bacteria [21]. These microorganisms are
particularly sensitive to changes in temperature and pH [19], their
development is inhibited by a high level of volatile fatty acids and
other compounds, that is, hydrogen, ammonia, sulphur hydrogen in
the environment [5].
The end product can be formed, either by means of cleavage of
acetic acid molecules to generate Carbon dioxide and methane or by
the reduction of Carbon dioxide with hydrogen to form methane and
water [20]. Besides the above two groups, some methane can also be
produced by the methylotrophic methanogens [5,,16,17].
There are three groups of methanogens, namely acetotrophic,
hydrogenotrophic, and methylotrophic [16]. The majority of the
methane is produced by acetotrophic methanogens, which transform
the acetate (resulting from acetogenesis) into methane and carbon
dioxide. The hydrogenotrophic group converts hydrogen and carbon
dioxide into methane [20].
74.5 % of archaea species utilize hydrogen and carbon dioxide, 33
% utilize methyl compounds and 8.5 % utilize acetate. The utilization
of methyl compounds (mainly micro-organisms belonging to the
genera Methanosarcina and Methanolobus) is seldom accompanied
by an ability to utilize hydrogen and carbon dioxide [34]. Some
members of acetoclastic methanogens include Methanosarcina,
Methanothrix, Methanosaeta, etc. Hydrogenotrophic methanogens
include Methanospirillum, Methanoculleus, Methanobrevibacter,
Methanocorpusculum, etc [18]. However, the Methanosarcina
species could employ both the acetoclastic and the hydrogenotrophic
methanogenesis pathways [20]. Methanogens are the most important microorganisms in the anaerobic digestion process converting
organic matter to methane.
Microbial Identification in Ad:
Identifying the species in a sample is crucial in microbiology
research. Most of the microorganisms involved in AD are anaerobes
and their cultivation in the laboratory is one of the most challenging
areas of microbial research [35].A. Culture-based techniques of Microbial Identification in AD:
Before the advent of molecular tools such as metagenomics, the
microbial ecology of various environments including those of the
anaerobic world was largely elusive [14]. The culture-dependent
methods have various limitations which lead to incomplete or even
incorrect information on microbes isolated. It is usually only a small
fraction of the microbes in anaerobic digestion plants that can be
cultured because the artificial growth media may not adequately
simulate the environment in the anaerobic digestion plants or
provide all the nutrients required for the growth of the microbes
[36]. Many microbes require syntrophic interactions with others, and
thus they cannot be cultured individually, they require co-culturing
of the microbes. Some microorganisms have similar physiological,
biochemical and/or morphological characteristics; therefore, they
cannot be distinguished from one another with certainty. As a result,
it has been estimated that only one per cent of the microorganisms
present in anaerobic environments had been isolated or characterized
[37].B. Molecular biology techniques of Microbial Identification in AD:
Only a few per cent of bacteria and archaea have so far been
isolated, but little is known about the dynamics and interactions
between these microorganisms. The lack of knowledge results
sometimes in malfunctions and unexplainable failures of biogas
fermenters. With molecular techniques, more information can be
received about the community structures in anaerobic processes [4].A variety of 16S rRNA-based techniques have been developed
and applied to microbial ecological studies of biogas-producing
microbiomes. Cloning of PCR-amplified 16S rRNA gene, a fragment
or the entire gene, followed by sequencing of individual clones with
the Sanger sequencing technology has been used for decades in the
analysis of microbiomes, including biogas-producing microbiomes
[36-38]. The recent advancements in next-generation sequencing
(NGS) technologies have made this traditional method obsolete.
Metagenomics, a culture-independent method allows for the
direct examination of microbial community structure and function
in an ecosystem using various bioinformatics pipelines [39]. The
application of omics-based studies has revealed a number of things
previously unknown to the anaerobic microbial world such as new
taxa and their roles in various anaerobic systems [39].
The methods of metagenomics provide extensive insight into
microbial phylogeny in AD [40]. Meta-omics techniques and gene
amplicon sequencing methods can fill this gap in the understanding
of AD and have been developed in order to link the function and
activity of the microbial community.
Metagenomic approaches are appropriate strategies for different objectives such as identifying and isolating key players of an
anaerobic culture [40]. Provide greater information than amplicon
gene sequencing approaches [13].
Molecular biology techniques provide valuable tools for improved
understanding of microbial communities and their function in
connection with different aspects of AD, which in turn may help
optimize the biogas production process more efficiently. A broad
range of studies was published recently on investigations of microbial
community structures in biogas reactors. The methodologies applied
included analysis of total bacteria and archaeal community by
targeting 16S rRNA using 454 next-generation sequencings (NGS)
technique; as well as detection and quantification of methanogenic
Archaea by quantitative real-time polymerase chain reaction (qPCR)
[41]. The traditional molecular biology technologies help with
identifying only the most abundant microbial populations present in
the reactor. Due to their high sequencing depth, the newly developed
sequencing techniques make the determination of both the most
abundant and also the minor populations possible. The NGS-based
metagenomic approach enables following up on changes in the
microbial community structure starting from the very initial stage to
the souring of the digester [42].
Investigations of the microbial community in 21 full-scale
anaerobic digestion plants using 454 pyrosequencing of 16S rRNA
gene sequences showed that the bacterial community was always
more abundant and more diverse than the archaeal community in
all reactors.
Similarly, the denaturing gradient gel electrophoresis (DGGE)
technique is still among the promising methods to perform a
preliminary analysis of the microbial community profile and monitor
the various experimental stages during the biogas production process
[43].
Moreover, the high-throughput Illumina Miseq approach is
also widely considered a promising culture-independent method
to perform microbial community analysis of AD systems. By the
application of this method, the specific syntrophic relationships
between acetogens and methanogens could be better understood,
especially in terms of how they can be related to disturbances
occurring in the biogas production process [44,45].
Conclusion
The role of microbes in anaerobic digestion cannot be
overemphasized. These tiny creatures use a combined effort to
transform different biomasses into valuable materials, maintaining
the ecosystem in a unique and equilibrium state. Microbes which are
ubiquitous can be harnessed to help man solve his numerous needs.
The microbiota involved in anaerobic digestion should therefore be
elaborately studied and exploited as more biomasses are used. These
microbes can be further bioengineered to breakdown xenobiotics in
anaerobic conditions to produce valuable materials. Unraveling the
synergy amongst the microbiota involved in AD will not only help in
the improvement and efficiency of the AD process but will also lead
to the optimization of biogas production.