Journal of Veterinary Science & Medicine
Download PDF
Research Article
Isolation and Multiple Drug Resistance Patterns of Salmonella Isolates from Selected Dairy Farms in Hawassa Town, Ethiopia
Fesseha H*, Aliye S, Kifle T and Mathewos M
School of Veterinary Medicine, Wolaita Sodo University, Ethiopia
*Address for correspondence: Haben Fesseha, School of Veterinary Medicine, Wolaita Sodo University,
Ethiopia, PO Box 138, Wolaita Sodo, Southern, Ethiopia; E-mail:
tseyon.h@gmail.com
Submission: 20-January-2020
Accepted: 21-February-2020
Published: 22- February-2020
Copyright: © 2020 Fesseha H Heredia Peralta DT, et al. This is an
open access article distributed under the Creative Commons Attribution
License, which permits unrestricted use, distribution, and reproduction in
any medium, provided the original work is properly cited.
Abstract
A cross-sectional study was conducted from November 2017
to May 2018 on selected dairy farms in Hawassa town to isolate and
assess the in-vitro antibiotic biogram of Salmonella from lactating dairy
cows, personnel’s and equipments at farms. A total of 216 samples
were collected from selected dairy farms. All samples were processed
bacteriologically following standard procedures outlined by ISO 6579:
2002. The overall prevalence of Salmonella was 12.9% (28/216) of the
total samples. Out of total, 64.3% (18/28), 10.7% (3/28) and 25% (7/28)
were from lactating cows, personnels’, and equipments, respectively.
Based on antimicrobial susceptibility testing, all isolates were resistant
at least to one or more antimicrobials tested. Accordingly, 96.4%
(27/28), 82.1% (23/28) and 75.0% (21/28) isolates showed resistance for
oxytetracycline, kanamycin, and nalidixic acid, respectively. Out of all
the resistant isolates, 96.4% (27/28) showed multiple antibiotic resistance
(resistance to two or more antibiotics) patterns. Multiple antimicrobials
resistance was observed in 66.7% (18/27), 7.4% (2/27) and 25.9% (7/27)
from lactating cows, personnels’, and equipments, respectively.
Thus, awareness creation to the public regarding the public health
importance of multiple drug-resistant Salmonella species and the
consumption of unpasteurized milk and milk products is important.
Keywords
Dairy farms; Isolation; Hawassa; Multiple drug;
Antimicrobial resistance; Salmonella
Introduction
Milk has been described as a nearly perfect food since it contains
the vital nutrients essential to the body, but it is also considered as
a good medium for many microorganisms [1]. Raw untreated milk
is still used by a large number of farm families and workers. In the
raw milk value chain, milk producers, vendors and shop outlets can
influence the prevalence of harmful pathogens in milk through poor
animal husbandry, adulteration, washing equipment, udder and
hands with unsafe water, storing and transportation in unhygienic
condition and abuse of storage temperature [3]. Especially, the safety
of dairy products with respect to foodborne diseases is a major global
issue especially in the developing countries where production of
milk and milk products takes place under poor hygienic, sanitary
and Agricultural practices [3]. Milk contamination by zoonotic
pathogenesis often natural but can also occur through handling milk
in unhygienic conditions [1,4].
Food-borne bacterial diseases are a serious challenge to public
health in developed and developing countries. There are more than
250 different food-borne diseases and most of these diseases are
infections, caused by a variety of bacteria, viruses, parasites, and
poisonings caused by harmful toxins or chemicals like poisonous
mushrooms [5,6]. There different bacteria that cause foodborne
diseases such as Salmonella, Campylobacter, Listeria, pathogenic
Escherichia coli, Yersinia, Shigella, and Enterobacter. Salmonella is one of the most important bacterial species that infect a wide variety of hosts including humans and numerous farm animals; such as pigs,
cattle, horses, and chickens [7].
Salmonella is comprised of different species and more than 2,600
different serovars of Salmonella have been characterized based on
the surface ‘O’ antigen, which is a part of the variable long chain
of lipopolysaccharide on the bacterial outer membrane [8]. Out of
these 2,600 serovars, nearly 1500 belong to the Salmonella subspecies
enterica. Serovars of the enterica subspecies can be divided into three
groups depending upon their ability to infect a wide variety of hosts.
The first group includes serovars that have a broad host range also
called unrestricted serovars as these infect nearly all animals. This
group includes serovars like Salmonella typhimurium and Salmonella
enteritidis. Nevertheless, these serovars are of high importance with
respect to their epidemiology as these have developed mechanisms
to invade different hosts without any greater resistance. Thus,
these serovars pose a greater zoonotic potential than their other
counterparts [9].
The second group includes serovars that cause highly severe
systemic infection in their preferred host and are usually excreted
without any clinical symptoms when they accidentally infect hosts
others than their most adapted or preferred. Serovars such as Dublin,
Choleraesuis fall into this category, as these prove to only cause
systemic infection in cattle and pigs respectively [10]; however, these
upon infection into other hosts like rodents and humans are usually
excreted making these hosts as ‘carriers’. Serovars of this group are
referred to as the ‘Host-adapted Serovars’. The third group comprises
of serovars which are restricted very strictly with one very specific
host only; these serovars are called ‘host restricted serovars’. They
exclusively cause systemic infection, which often proves to be fatal
within their host. Serovars such as Typhi, Gallinarum, Abortus equi,
ectecra belong to this group [11].
Salmonella is transmitted to animals and humans through
consumption of contaminated food products (milk, eggs, and
meats), direct contact with infected animals, through contaminated
equipments such as stainless steel, hanging material, bucket, where
milk is collected and stored, are a key mechanism for pathogens to
contaminate food products [12]. In livestock, clinical signs typically
appear 6-24 hr after exposure and include profuse diarrhea, fever, dehydration, in appetence, foul-smelling feces, and mucus or blood in feces [13]. Disease manifestations in people include diarrhea, fever, abdominal cramps and septicemia in severe cases, appearing 12-72 hr
after ingestion. Salmonella can also be carried subclinically by both
humans and animals [14,15].
The prevalence of salmonella infection varies across regions,
however, the diseases caused by S. enteric serovars are especially
prevalent in developing areas, such as Southeast Asia, Africa and
South America that leads to an estimated 20 million cases of humans
and 200,000 deaths each year. Challenges such as antibiotic-resistant
Salmonella strains also pose a significant threat to deliver reliable
therapies [16]. In Ethiopia, as in other developing countries, it is
difficult to evaluate the burden of Salmonellosis because of the
limited scope of studies and the lack of coordinated epidemiological
surveillance systems. In addition, under-reporting of cases and the
presence of other diseases considered to be of high priority may have
overshadowed the problem of Salmonellosis. Continuous surveillance
of the prevalent Salmonella serovars and assessing their antimicrobial
resistance pattern is essential to control the spread of the pathogen
[17].
Antibiotic resistance in Salmonella is a rising problem over
the past decades. Improper use of antibiotics in both human and
veterinary medication has caused bacteria to develop resistance
against therapeutic antibiotics [18,19]. Using antimicrobial agents
for cattle has been implicated as a source of human infection with
Antimicrobial-Resistant (AMR) Salmonella through direct contact
with livestock and consumption of raw milk, meat and contaminated
material [20]. Antimicrobial-resistant Salmonella are increasing due
to the use of antimicrobial agents in food animals at subtherapeutic
level or prophylactic doses that may promote growth and markedly
increase the human health risks associated with consumption of
contaminated milk and meat products through mutation, acquisition
of resistance encoding genes and irrational use of antimicrobials in
food animals [21-23].
Accordingly, there are limited studies regarding the assessment of
the pathogens isolated from apparently healthy animals at farm level,
personnels’ and different types of equipment. Thus, the screening of
milk and other dairy products against pathogenic organisms will play
a vital role in curtailing human infection. Studying the prevalence and
antimicrobial resistance of Salmonella from cattle and in contact with
humans in dairy farms is the most important to design methods of
minimizing the possible transmission of Salmonella between humans
and cattle. Moreover, it important in combating the emergence of
antibiotic-resistant strains of Salmonella [11]. Hence, this study was
conducted to isolate, identify and assess the multiple drug resistance
pattern of Salmonella isolates from selected dairy farms in Hawassa
town.
Materials and Methods
Study area: The current study was conducted from November 2017 to May
2018 in selected dairy cattle farms of Hawassa towns. It is located 275
km south of Addis Ababa. Hawassa is situated at an altitude of 1750 m
above sea level and according to an estimate, it lies between 6 °83’ to
7 °17’ N and 38 °24’ to 38 °72’ E. Hawassa receives an average annual rainfall of 955 mm with mean annual temperature of 20 °C and the
city has a total area of about 50 km2 divided into eight sub-cities and
32 kebeles (kebeles are the smallest administrative unit below the subcity/
woreda level) [24].
Study population: The study animals were apparently healthy dairy cows that were
located in Hawassa town. The study includes dairy cattle kept under
different (extensive, intensive and semi-intensive) management
systems as well as farm personnels and equipments. There are
different types of farms including small, medium and large scale
having dairy cattle ranging from five to twenty two. Besides, the farms
were selected purposively based on the availability of lactating cows
and the willingness of the owners.
Study design and Sampling technique: A cross-sectional study was carried out from November 2017
to May 2018 to isolate, identify and assess the multi-drug resistance
pattern of the salmonella isolates from selected dairy farms. The farms
are selected purposively based on the availability and accessibility of
study animals. Accordingly, a total of 216 samples were collected
from selected dairy cattle farms in the study area.
Sample collection, handling and transportation: Samples were collected aseptically from apparently dairy cows
(milk and feces), hands of personnel working in the farms and
from equipment (container and buckets). Then the all samples
were collected after getting proper consent from the personnel and
Hawassa university to perform the research activity. Fecal samples
were collected directly from the rectum and put into 50 ml containing
a universal screwed capped bottle and 10 ml of milk was collected
aseptically from all teats in a sterile test tube after aseptically
preparing the teats thorough scrubbing with a cotton moistened
with 70% denatured alcohol and the first 3-4 streams of milk were
discarded. All types of swab samples (milkers’ hand, container and
buckets) were collected before the commencement of the milking
process using a sterile wooden cotton swab and were put into a sterile
test tube containing 10 ml buffered peptone water used as transport
media. All sample types were properly labeled with permanent
marker. Then, the samples were immediately transported using an
icebox to the Microbiology Laboratory of Hawassa University for
further bacteriological examination.
Isolation and identification of salmonella: The isolation and identification of Salmonella was performed at the
Microbiology laboratory of Hawassa University by using techniques
recommended by International Organizations for Standardization
(ISO-6579, 2002) [25]. The detection of salmonella was performed
based on the following four successive stages: Firstly, All samples were
pre-enriched in non-selective liquid media and processed separately.
Then, 1 gm of fecal sample and 1 ml of milk was pre-enriched with
9 ml of Buffered Peptone Water (BPW) and incubated for 24 hrs
at 37 °C. Secondly, all samples were transferred to selective media
such as Tetrathionate Broth and Rappaport Vassiliadis Salmonella
Enrichment Broth. A 1 ml of pre-enriched sample was transferred
aseptically into a tube containing 10 ml of Tetrathionate Broth and
incubated at 37±1 ºC for 24±3 hours. Another 0.1 ml of the culture obtained in pre-enrichment broth was transferred aseptically into a
tube containing 10 ml Rappaport Vassiliadis Salmonella Enrichment
Broth (Harmonized) and incubated at 41.5±1 ºC for 24±3 hrs.
Thirdly, Plating out and identification of the samples were
conducted using Xylose lysine Desoxycholate (XLD) agar and
Salmonella-Shigella (SS) agar plates. A loopful of inoculum from
cultures of the selective enrichment media were streaked on to
XLD and SS agar plates and incubated at 37 ºC for 24 hrs. Then, all
colonies that grow on the XLD medium, produces hydrogen sulfide
(H2S) and colorless colonies with black center on SS medium were
streaked onto Nutrient Agar and incubated at 37 ºC for 24 hrs for
further confirmation through serious of biochemical tests. Finally, All
suspected colonies were subjected to a series of different biochemical
tests using the procedure of (ISO 6579, 2002; to confirm salmonella
[25]. Triple Sugar Iron Agar (TSIA), Urease, Citrate, Indole, Methyl
red and Voges Proskouer (VP) tests were performed on all suspected
isolates to confirm the salmonella. All presumptive salmonella
Isolate were cultured on Nutrient Agar for further antimicrobial
susceptibility testing.
Antimicrobial susceptibility test: The antibiotic susceptibility tests of the Salmonella isolates were
performed according to the Clinical and Laboratory Standards
Institute (CLSI) method using Kirby-Bauer disk diffusion test on
Muller-Hinton Agar (HIMEDIA, India) [26]. Pure colonies on
nutrient agar were taken with a wire loop and transferred to a tube containing 5 ml of Saline water and emulsified. The broth culture
was incubated at 37 °C for 4 hrs until it achieved the 0.5 McFarland
turbidity standards. Sterile cotton swab was dipped into the suspension
and the bacteria were swabbed uniformly over the surface of Muller-
Hinton agar plate within a sterile safety cabinet. The plates were held
at room temperature for 15 minutes to allow drying. Antibiotic discs
with a known concentration of antimicrobials were placed and the
plates were incubated for 24 hrs at 37 °C.
Amoxicillin (AML) (25 μg), Cefoxitin (FOX) (30 μg),
Chloramphenicol (C) (30 μg), Gentamycin (CN) (10 μg),
Streptomycin (S) (10 μg), Kanamycin (K) (30 μg), Nalidixic acid (NA)
(30 μg), Ciprofloxacin (CIP) (5 μg), Oxytetracycline (OT) (30 μg)
and Trimethoprim-sulphamethoxazole (SXT) (25 μg), were selected
based on availability and their current use in human and veterinary
medicine. All the antibiotics were from Oxiod Hampshire, England,
and the expiry date was properly checked before application. Zone
of inhibition of individual antibiotic agent was interpreted in to
susceptible, intermediate, and resistance categories by referring
recommended clinical and laboratory standards institute [26].
Data analysis: Data collected from field and laboratory investigations were
recorded, and coded using Microsoft Excel 2013 program and
analyzed using STATA version 13.0. Descriptive statistics were used
to figure out the proportions of Salmonella isolate. Moreover, the
antibiotic efficacy of each drug was determined by comparing the zone of inhibition with the standard one.
Results
Frequency of Salmonella isolates: In this study, out of 216, the overall prevalence of Salmonella was
12.9% (28/216). From the overall proportion, 64.3% (18/28), 10.7%
(3/28) and 25% (7/28) were isolated from the milk and feces of dairy
cows, personnel and equipments, respectively. 19% of Salmonella
were isolated from plastic container milk. A higher proportion of
Salmonella was isolated from milk samples (12.1%) than fecal samples
(7.7 %) (Figure 1).
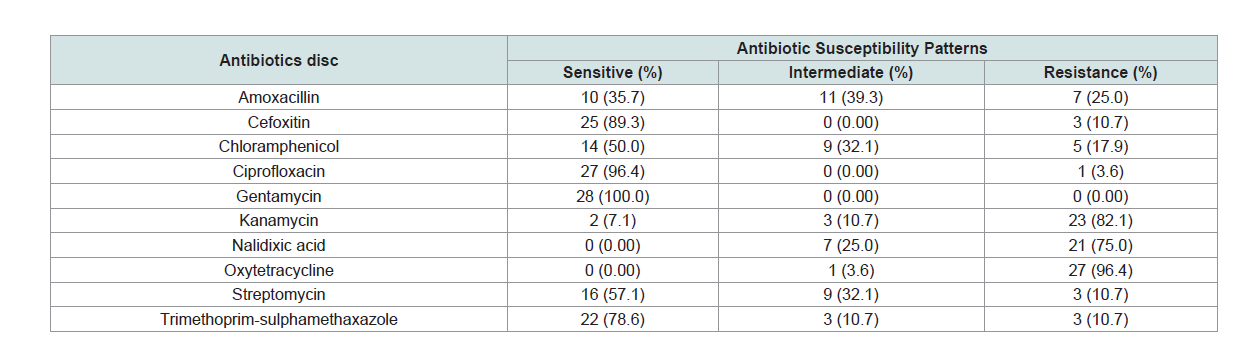
Antimicrobial susceptibility of Salmonella isolates: In the present study, out of 28 isolates, 27 isolates showed
multiple drug resistance. Accordingly, all isolates were susceptible to
ciprofloxacin, cefoxitin and trimethoprim-sulphamethoxazole with
proportion of 96.4%, 89.3%, and 78.6%, respectively. However, all
isolates were 96.4%, 82.1% and 75.0% resistant to oxytetracycline,
kanamycin and nalidixic acid, respectively. On the other hand, all
isolates were 100% sensitive to gentamycin (Table 1).
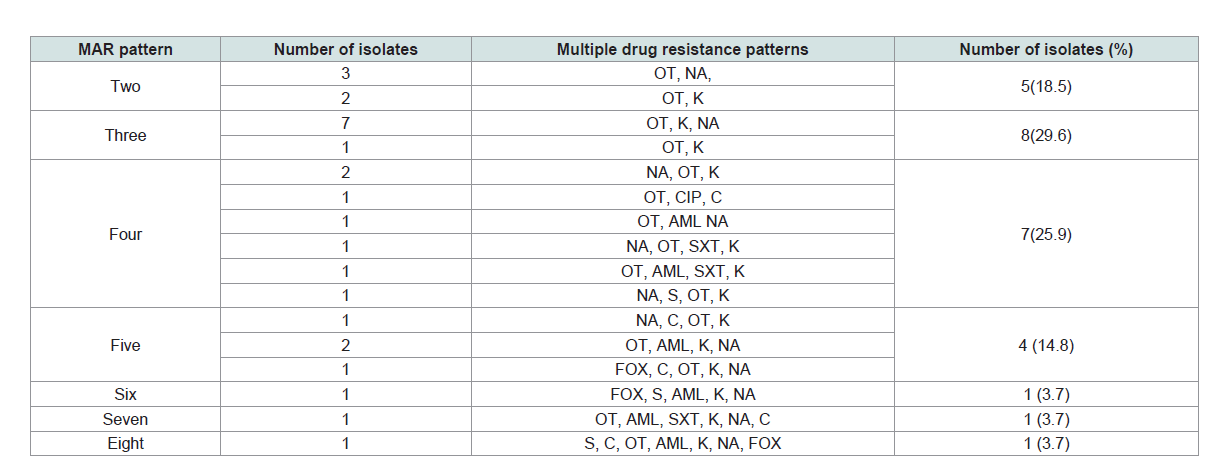
Multiple drug resistance patterns of Salmonella isolates: Multiple drug resistance (isolates that were resistant for two or
more antibiotics) were detected in 96.4% (27/28) of the Salmonella
isolates. Out of these, 66.7% (18/27), 7.4% (2/27) and 25.9% (7/27)
isolates were from lactating cows, personnels’, and equipments,
respectively. The higher multi-drug resistance pattern was observed
in K, NA, OT, with the proportion of 25.9% followed by K, NA, OT,
AML with the proportion of 7.4%. Besides, 11.1% of the resistant
isolates were resistant to six and more antibiotics (Table 2).
Discussion
In this study, out of 216 samples collected from selected dairy
farms in Hawassa town, the overall proportion of Salmonella isolated
from dairy cows, personnels’ and equipment were 12.9%. This was
higher than the reports of where 7.2% were found in slaughtered
small ruminants and environment in Modjo export abattoir [27],
7.1% from apparently healthy slaughtered cattle in Debre Zeit and the
study on cheese and milk in Debre Zeit (2.1%) as well as dairy product in Addis Ababa (1.6%) [21,28-30]. However, the current finding
was comparable with 10.5% from apparently healthy dairy cows in
Modjo [31], 10.76% from lactating cows and in contact humans in
dairy farms of Addis Ababa and (11.5%) among chicken table eggs at
Kombolcha [18], Ethiopia [32]. The present result was lower than the
findings of who reported 20% in raw milk from the Korsa district and
Ejeta et al., 2004 who reported 14.7% from minced beef, mutton and
pork samples among supermarkets in Addis Ababa [33].
In this study, the prevalence of Salmonella from milk and feces
of apparently healthy lactating dairy cows was 64.3%. This was
higher than who reported 7.1% from apparently healthy slaughtered
cattle [28]. This variation could be due to the test procedures and
techniques used since pre-enrichment steps using buffered peptone
water was employed in this study and source of sample. Similarly, the
report of [34]; from England (0.2% and 4%), from Northern Thailand
(3%) and from Cameroon (27%) are much lower than the current
study [35,36]. The current result was higher than the prevalence
recorded in Iran 4% and in USA 7.3% and in Nigeria (15%) and 10.9%
reported in Namibia on bovine and ovine bone-and-meat meal and
blood meal samples [19,37,38]. This may be attributed to the variation in agroecological location of the cattle, housing conditions, feeding
habits, and types of feed provided for the cattle.
According to the current investigation, Salmonella was isolated
from the fecal samples of apparently healthy lactating dairy cows
with a rate of 7.7%. This finding was higher than the report of from
Egypt where prevalence in on fecal shedding of Salmonella among
dairy cattle was 1.56 [39]. However, this result was lower than from
the United States (9.7%) [40], from central Texas [41], USA where
Salmonella shedding rate from fecal samples of dairy calf was 70%.
This huge difference might be in the report from Texas, all isolates
were one serotype (S. kinshasa) and this serotype might have specific
host requirement.
In the present study, Salmonella was isolated from milkers’ hand
swab with a rate of 14.3%. This was higher than the report of (8.9%)
from small ruminants slaughtered in Modjo export abattoir [27]However, it was lower than the work of Beyene et al., 2016 (28.6%)
from pooled milkers’ hand swab of personnels’ working in Asella
Municipal abattoir.
The variation in the prevalence of Salmonella isolation between
the present-day study and the previous studies at different areas of the
country could be associated with different risk factors that contribute
to the occurrence of Salmonella. These are host-related risk factors
that include age, breed, the physiological state of the animals, feeding
strategies, vaccination status [29]. Environment-related risk factors
are often related to hygienic and management practice, stocking
density, type and amounts of feed, accessible water supplies, infection
load in the environment, usage of contaminated utensil, housing type,
ventilation, flooded grassing areas, movement of animals, calving
environment, and production facilities in different areas are also plays
a role for Salmonella occurrence [12]. Additionally, epidemiological
patterns of Salmonella differ greatly between geographical areas
depending on climate, population density, land use, farming practice,
food harvesting and processing technologies and consumer habits
[42].
The current study revealed that 96.4% of the isolates were resistant
for two or more antibiotics which was comparable with the finding
of [31]. However, it was higher than the previous studies conducted
in Ethiopia and elsewhere in the world [21,28,43-46]. This difference may be due to the increasing rate of inappropriate utilization of
antibiotics in the dairy farms which favors selection pressure that increased the advantage of maintaining resistance genes in bacteria [47,48].
The result of the current research indicated Salmonella isolates
were resistant to Oxytetracycline, kanamycin, and nalidixic acid
with a resistance rate of 96.4%, 82.1%, and 75% respectively.
Similarly, reported that the isolates of Salmonella from food items
and personnel from Addis Ababa were resistant to the commonly
used antibiotics including streptomycin [21], and oxytetracycline.
However, resistance rates to oxytetracycline are very high compared to
results documented in America reported 95.6% and 87.8% sensitivity
levels [37], respectively and Iran reported 42.58% sensitivity for both
antibiotics [19]. In this study, 96.4% of the isolates showed resistance
to two or more antibiotics which is lower than a report from Addis
Ababa, Ethiopia (83%) [18].
According to the study, Salmonella isolates were susceptible to
gentamycin and ciprofloxacin with the rate of susceptibility 100% and
96.4% respectively. This was in agreement with the reports of where
Salmonella isolated from apparently healthy slaughtered sheep in
Turkey showed 100% sensitivity to these antibiotics [36], with and
with the report of in Iran where ciprofloxacin was 100% effective
[19,30]. However, it was higher than who reported 73.3% and 83.3%
[11], who reported 75% and 95% for ciprofloxacin and gentamycin,
respectively [33]. This variation might be due to small sample sizes
for the data, nature of the drug, presence of different strains of the
bacteria, development of resistant gene, their low-frequency usage for
prevention and control of disease in food animals in the study area.
The present study revealed that Salmonella isolates were
resistant to tetracycline and ampicillin with a rate of 96.4% and 39%,
respectively which disagrees with the report of in Egypt reported that
each of the ampicillin and tetracycline was 85.7% effective against
Salmonella species isolated in dairy cattle [39]. In addition, in the
present study trimethoprim-sulphamethoxazole was an effective drug
(78.6%) against salmonella isolates that disagrees with the report by who reported 100% resistance to trimethoprim-sulphamethoxazole
[39]. A higher activities of gentamycin (100%) observed in the current
study disagree with a study in Texas, USA, reported 85% and this
difference might be due to availability and overuse of the drug in the
farm of the current study [41]. In the current study, ciprofloxacin
was 96.4% effective against all isolates which was in line with a report
in Sudan where ciprofloxacin was 100% effective to all human and
cattle Salmonella isolates [49]. The result for streptomycin resistance
in this study (10.7%) was lower than 13.3% and 25%, which was
reported by and [18,33], respectively. Amoxicillin resistance in this
study (25%) was higher than 16.7% reported by [30]. The resistance
of chloramphenicol in this study 17.9% is consistent with 16.7% reported by and [18,30], and lower than 25% reported by [33].
According to the antimicrobial susceptibility testing, all of the
isolates showed multiple drug resistance to at least one or more drugs
tested were observed which was in line with the report of [30,33,50]. Moreover, 96.4% of the isolates showed multiple drug resistance for
two or more types of antimicrobials. This was higher as compared to
the report of who reported 70% and 30% [33], who report 83.3% and
16.3% [50], and who reported 50% and 50% for multiple and single
antimicrobial resistance, respectively [30].
In general, antimicrobial use is a key driver of resistance
development, which is either overuse for minor infectious, misuse
due to lack of access to appropriate treatment and underuse due to
inadequate dosing, poor adherence or substandard antimicrobial and
lack of financial support to complete treatment course. The present
study indicated the importance of cattle products (milk), personnel
working in the farms and materials/equipment used as a potential
source of Salmonella infection.
Conclusion and Recommendations
In the present study, the isolation of 12.9% Salmonella at dairy
farms level showed that dairy cattle and their environment are
important sources of milk contamination with the organism, and
consumption of raw milk and other unpasteurized dairy products
can lead to infection with zoonotic Salmonellosis. The presence of a
high proportion of multiple antimicrobial-resistant isolates (96.4%)
in the dairy farms to antimicrobials that are commonly used in the
veterinary and public health set up in this study further signifies the
public health importance of Salmonella in addition to treatment
failure. In this study, all the isolated Salmonella revealed resistance at
least to one of the antibiotics tested. In general, awareness creation to
the public about the public health importance of foodborne diseases
and the consumption of unpasteurized milk and milk products is
important. Gentamycin and Ciprofloxacin should still be used as a
choice to treat Salmonellosis. Further, the molecular characterization
of the isolates with emphasis on resistant strains is important to
identify mechanisms of antibiotic resistance.