Journal of Microbiology & Microbial Technology
Download PDF
Research Article
*Address for Correspondence: Ligimol James, Department of Dairy Microbiology, College of Dairy Science and Technology, Kerala Veterinary and Animal Sciences University, Mannuthy, Thrissur, Kerala, India, E-mail: lijimoljames@kvasu.ac.in
Citation: James L, Beena AK, Anupa A, Sreeshma N. Antibiogram of Lactobacilli Isolated from Four Different Niches. J Microbiol Microb Technol 2016;1(1): 4.
Copyright © 2016 James L, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Microbiology & Microbial Technology | Volume: 1, Issue: 1
Submission: 25 May, 2016 | Accepted: 30 May, 2016 | Published: 03 June, 2016
Reviewed & Approved by: Dr. Mohamed Aleraky Saleh, Clinical and Medical Microbiology, Al-Azhar University, Egypt
Antibiogram of Lactobacilli Isolated from Four Different Niches
Ligimol James*, Beena AK, Anupa A and Sreeshma N
- Department of Dairy Microbiology, College of Dairy Science and Technology, Kerala Veterinary and Animal Sciences University, India
*Address for Correspondence: Ligimol James, Department of Dairy Microbiology, College of Dairy Science and Technology, Kerala Veterinary and Animal Sciences University, Mannuthy, Thrissur, Kerala, India, E-mail: lijimoljames@kvasu.ac.in
Citation: James L, Beena AK, Anupa A, Sreeshma N. Antibiogram of Lactobacilli Isolated from Four Different Niches. J Microbiol Microb Technol 2016;1(1): 4.
Copyright © 2016 James L, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Microbiology & Microbial Technology | Volume: 1, Issue: 1
Submission: 25 May, 2016 | Accepted: 30 May, 2016 | Published: 03 June, 2016
Reviewed & Approved by: Dr. Mohamed Aleraky Saleh, Clinical and Medical Microbiology, Al-Azhar University, Egypt
Abstract
Presence of antibiotic resistance in lactobacilli, invites special attention from the public health point of view as they are widely used as starter cultures and probiotics. Lactobacillus, a ubiquitous genus, is expected to have an antibiogram that vary remarkably with their source. In the present work antibiotic resistance/sensitivity of Lactobacillus species isolated from four different sources: carrot, idli batter, curd and duck faeces were assessed by the Disc diffusion assay. Contrary to the earlier reports of general sensitivity of lactobacilli to the inhibitors of cell wall synthesis, all the four Lactobacillus isolates were found to be resistant against the cell wall synthesis inhibitors ampicillin and cephalosporins (cefalexin and cefixime) irrespective of their source of isolation. However the isolates were found to be sensitive to protein synthesis inhibitors like azithromycin, tetracycline, gentamicin, clindamycin and chloramphenicol. In general the isolates were found to be more resistant to those antibiotics that act by inhibiting cell wall synthesis than those which act by inhibiting the protein synthesis. This observation of multidrug resistance by the lactobacilli isolated from different niches reemphasize the significance of considering the antibiogram study as a critical criteria while screening lactobacilli isolates for use in food formulations. Molecular level studies are indicated to have an in-depth understanding of the basis of resistance.Keywords
Lactobacilli; Antibiotic resistance; Cell wall inhibitors; Protein synthesis inhibitorsIntroduction
Lactic acid bacteria (LAB) are Gram-positive, catalase negative, acid -tolerant and non-spore forming cocci and rods. The common feature of the ability of LAB to produce lactic acid as a major end product of fermentation of hexoses makes them the potential agents for most of the food fermentations. The LAB are present in a number of natural habitats and are widely utilized as starter cultures and probiotics. Among LAB, The genus Lactobacillus is the largest group comprising around 140 species and 30 subspecies [1,2]. Though Lactobacilli have a long history of safe use in the production of fermented foods and beverages, recent reports that indicate that the commensal bacteria including lactic acid bacteria (LAB) could act as reservoirs of antibiotic resistance genes raises concern about the safety of their use [3.4]. Considering the reports of possibility of horizontal transfer of resistance genes between bacterial species, research on the antibiotic resistance of lactobacilli has gained much momentum nowadays. Lactobacilli are intentionally added to food for carrying out desirable fermentations and in the case of probiotic products they should essentially retain their viability during passage through the GIT (Gastrointestinal Tract). So the chances are high that they get ample opportunities to act as potential reservoirs for the transfer of antibiotic resistance genes to pathogenic bacteria. Hence safety evaluation of Lactobacillus isolates is highly critical to ensure consumer well-being. Such an assessment also helps in identifying lactobacilli that could be advised for replenishing gastrointestinal microflora during antibiotic therapy. Information in this regard will also be helpful in designing media with selective properties for the isolation of specific lactobacilli from mixed bacterial populations. Also considering the possibility that their natural habitats might be having a profound influence on their antibiotic resistance pattern, this study was designed to conduct the safety assessment of four lactobacilli isolates from different sources and maintained in the culture collection of Dairy Microbiology Department, College of Dairy Science and Technology, Mannuthy, Thrissur, Kerala, through antibiotic-susceptibility assays.Materials and Methods
Lactobacilli culturesFour Lactobacillus isolates maintained in the stock culture collection of Department of Diary Microbiology, College of Dairy Science and Technology were used for this study. The isolates (1,2,3 and 4) were from carrot, idli batter, curd and duck faeces respectively. The cultures preserved as glycerol stock were activated by growing in sterile MRS (deMan, Rogosa, Sharpe, Himedia, Mumbai) broth at 37 °C for 24 h. The cultures thus activated were stored under refrigeration with fortnightly activation in sterilized MRS broth. Working cultures were prepared by inoculating 0.1 ml of the stock culture in sterilized MRS broth followed by incubation at 37 °C for 48 hours.
Antibiogram of isolate
The antibiotic susceptibility of the isolate was determined as per the standard method [5]. Activated culture of each isolate was swabbed over the surface of Mueller Hinton agar (Himedia) agar plates. Fourteen different antibiotic discs (Amoxicillin (AM, 10 mcg), Ampicillin (A, 10 mcg), Azithromycin (AZM, 15 mcg), Aztreonam (AT, 50 mcg), Bacitracin (B, 10 units), Cefalexin (CN, 30 mcg), Cefixime (CFX, 5 mcg), Chloramphenicol (C, 30 mcg), Ciprofloxacin (CIP, 10 mcg), Clindamycin (CD, 10 mcg), Gentamicin (G, 50 mcg), Oxacillin (OX, 5 mcg), Sulphadiazine (SZ, 100 mcg), Tetracycline (T, 10 mcg), HiMedia Laboratories Pvt. Ltd, India) were placed over the surface of the inoculated plate. The plates were incubated at 37 °C for 24 h. The diameter of the Zone of inhibition around each disc was measured and expressed in mm. Results were interpreted as sensitive, S (≥ 21 mm); intermediate, I (16-20 mm) or resistant, R (≤ 15 mm) [6].
Results and Discussion
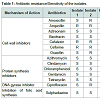
Irrespective of the source of isolation all the isolates exhibited remarkable resistance to a number of antibiotics revealing multidrug resistance (Figure 1). Among the isolates, those from idli batter and duck faeces were resistant against highest number of antibiotics (6/14).Of the 14 antibiotics tested eight antibiotics namely azithromycin, aztreonam, bacitracin, chloramphenicol, clindamycin, gentamycin, sulphadiazine and tetracycline were found to inhibit all the isolates used in this study (Figure 2). Mueller Hinton agars showing the zone of inhibition of different antibiotics to the tested isolates are shown in Figure 3. In general antibiotics elicit their action by inhibiting synthesis of bacterial cell wall, protein, folate or by inhibiting the action of DNA gyrase. Contrary to the earlier reports of sensitivity of lactobacilli to cell wall inhibitors all the four Lactobacillus isolates tested in this study were found to be resistant to half of the cell wall synthesis inhibiting antibiotics tested (Ampicillin, Cefalexin and Cefixime, Table 1 and Figure 4) [7]. Widespread sensitivity toward penicillins has already been reported in lactobacilli used as starter cultures [8]. Contrary to this, a pattern of resistance against Betalactams (Amoxicillin, Ampicillin and Oxacillin) was exhibited by the Lactobacillus isolates in the present study. Resistance exhibited by all the isolates against cephalosporin antibiotics (cefalexin and cefixime) is in agreement with the earlier reports of resistance of lactobacilli to cephalosporins [7]. Sensitivity was shown by all the isolates to the cell wall synthesis inhibitors, Bacitracin (targets cell membrane) and ztreonam. Variations were observed among the isolates in their response to the other two cell wall inhibitors tested namely, amoxicillin and oxacillin. Isolate 1 was sensitive to amoxicillin whereas all other isolates were resistant. In the case of oxacillin, isolates 2 and 4 were resistant to it and others were sensitive. Similar pattern was observed in the case of the DNA gyrase inhibitor, Ciprofloxacin with isolates 2 and 4 being resistant and others sensitive. All the isolates irrespective of their source of isolation were found to be sensitive to the protein synthesis inhibitors (azithromycin, chloramphenicol, clindamycin, gentamicin and tetracycline, Table 1 and Figure 5). Lactobacilli are reported to be highly resistant to aminoglycosides (gentamycin, kanamycin, streptomycin) that act by inhibiting protein synthesis [9]. Interestingly in this work, contrary to this report all the tested isolates were found to be sensitive to gentamycin, an aminoglycoside. Sensitivity exhibited by the isolates against the antibiotics, chloramphenicol, tetracycline and clindamycin is in agreement with earlier reports of susceptibility of lactobacilli to even low concentrations of many protein synthesis inhibitors including these antibiotics [9]. Sensitivity of the lactobacilli isolates to tetracycline is reported by other reseachers also [7,10]. However contradictory to this, resistance to tetracycline is reported as the most frequent among lactobacilli [11,12]. Earlier reports by a number of authors [8,13-15] ranking the resistance genes, tet(M) for tetracycline resistance as most common in LAB also supports the prevalence of tetracycline resistance in LAB. However in the present study all the isolates were found to be sensitive to this particular antibiotic.
Conclusion
Multiple antibiotic resistance in lactobacilli do raise an alarm about their safety aspects. However the fact that many LAB exhibits intrinsic resistance to antibiotics and that in many cases the resistance is of non- transmissible type reduces the severity/ intensity of this problem to some extent. Yet it is necessary to subject these phenotypically identified antibiotic resistant isolates to further molecular level studies to characterize the resistance as either transmissible or non-transmissible. As presence of intrinsic resistance as well as resistance due to chromosomal mutations poses a low risk of horizontal dissemination, the food industry always prefers to use strains with such attributes than those harbor highly transmissible antibiotic resistance genes. So it is always preferable to include the antibiotic resistance assessment as a selection criterion while screening for potential starter cultures and probiotics.References
- Bernardeau M, Vernoux JP, Henri-Dubernet S, Gueguen M (2008) Safety assessment of dairy microorganisms: the Lactobacillus genus. Int J Food Microbiol 126: 278-285.
- Claesson MJ, van Sinderen D, O'Toole PW (2008) Lactobacillus phylogenomics--towards a reclassification of the genus. Int J Syst Evol Microbiol 58: 2945-2954.
- Ashraf R, Shah NP (2011) Antibiotic resistance of probiotic organisms and safety of probiotic dairy products. Int Food Res J 18: 837.
- Devirgiliis C, Zinno P, Perozzi G (2013) Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front Microbiol 4: 301.
- Bauer AW, Perry DM, Kirby WM (1959) Single-disc antibiotic-sensitivity testing of Staphylococci; an analysis of technique and results. AMA Arch Intern Med 104: 208-216.
- Vlková E, Rada V, Popelářová P, Trojanová I, Killer J (2006) Antimicrobial susceptibility of bifidobacteria isolated from gastrointestinal tract of calves. Livest Sci 105: 253-259.
- Ammor MS, Flórez AB, Mayo B (2007) Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol 24: 559-570.
- Danielsen M (2002) Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48: 98-103.
- Gueimonde M, Sánchez B, G de Los Reyes-Gavilán C, Margolles A (2013) Antibiotic resistance in probiotic bacteria . Front Microbiol 4: 202.
- Belletti N, Gatti M, Bottari B, Neviani E, Tabanelli G, et al. (2009) Antibiotic resistance of lactobacilli isolated from two italian hard cheeses. J Food Prot 72: 2162-2169.
- Roberts MC (2005) Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245: 195-203.
- Korhonen JM, Danielsen M, Mayo B, Egervärn M, Axelsson L, et al. (2008) Antimicrobial susceptibility and proposed microbiological cut-off values of lactobacilli by phenotypic determination. Int J Probiotics Prebiotics 3: p257-p268.
- Lin CF, Fung ZF, Wu CL, Chung TC (1996) Molecular characterization of a plasmid-borne (pTC82) chloramphenicol resistance determinant (cat-TC) from Lactobacillus reuteri G4. Plasmid 36: 116-124.
- Gevers D, Huys G, Swings J (2003) In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol Lett 225: 125-130.
- Cataloluk O, Gogebakan B (2004) Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol Lett 236: 7-12.